library(tidyverse)
library(data.table)Manipulating Large(ish) Datasets with data.table
Learning Objectives
- Reading large datasets into R.
data.tablesyntax for manipulating data frames.- A data.table and dplyr tour
- Introduction to data.table.
- Efficient reshaping using data.tables
- Interactive Cheat Sheet
Data Frames and Motivation
Big data: Can’t read data into memory. Need to do distributed computing (like Spark).
Biggish data: Can read data into memory, but it’s very slow to manipulate it.
The data.table package helps you with biggish data.
data.frames are a base R class. We’ve learned about them before.There are many competitors to
data.frames:tibble,DataFrame(from Bioconductor), anddata.table.Julia (another statistical programming language) has its own
DataFramesclass. Python’s data frame packages arepandasandpolars.data.tableis among the fastest in most benchmarks: https://h2oai.github.io/db-benchmark/.- Python’s
polars, and Julia’sDataFrames.jlcome close and are faster in some benchmarks. dplyrdoesn’t come close.
- Python’s
So why did we learn the tidyverse?
- The Posit folks are better at marketing, so
dplyris more widely used. - For datasets with fewer than, like, one million observations, dplyr works fast enough.
- The Posit folks are better at marketing, so
Syntax
Reading in and Printing Data
Load the data.table package and (to compare) the tidyverse into R:
We’ll demonstrate most methods with the data from “NYC-flights14” dataset from the data.table package vignettes. I’ve posted it on my own webpage: https://data-science-master.github.io/lectures/data/flights14.csv.
Read in data with
fread()(for “file read”). It accepts all delimiters, which you (optionally) specify with thesepargument. The default is to guess the delimiter.flights <- fread("../data/flights14.csv")fread()will return adata.tableobject.class(flights)[1] "data.table" "data.frame"Use
fwrite()to write adata.tableobject to a file.Compare to
read_csv()in the tidyverse.flights_tib <- read_csv("../data/flights14.csv")read_csv()will return a tibble.class(flights_tib)[1] "spec_tbl_df" "tbl_df" "tbl" "data.frame"tibbles anddata.tables both print better thandata.frames.flights flights_tibYou usually use the base function
str()(for “structure”) to look at thedata.tableentries.## data.table way str(flights) ## Similar tidyverse way glimpse(flights_tib)You can use
as.data.table()to convert atibbleor adata.frameinto adata.table. But there is rarely a time when you’d do this, since you use data.table for large datasets that you read in withfread().temp_dt <- as.data.table(flights_tib) class(temp_dt)[1] "data.table" "data.frame"
Filtering/Arranging Rows (Observations)
Just like in the tidyverse, we use logicals to filter based on rows. The syntax for this is to place the logicals inside a bracket. Let’s find all flights that left JFK and arrived at LAX.
## data.table way flights[origin == "JFK" & dest == "LAX"] ## equivalent tidyverse way flights_tib %>% filter(origin == "JFK", dest == "LAX")To get a specific row, insert a number into the brackets.
## data.table way flights[c(1, 3, 207)] ## equivalent tidyverse way flights_tib %>% slice(1, 3, 207)Reorder rows by using the
order()function inside the brackets. Let’s reorder the rows alphabetically by origin, and break ties in reverse alphabetical order by destination.## data.table way flights[order(origin, -dest)] ## equivalent tidyverse way flights_tib %>% arrange(origin, desc(dest))Exercise: Use both data.table and the tidyverse to select all flights from EWR and LGA, and arrange the flights in decreasing order of departure delay.
Selecting Columns (Variables)
To select a variable (a column), you also use bracket notation, but you place a comma before you select the columns. This idea is that you are selecting all rows (empty space before comma).
There are lots of ways to select columns that keeps the new object a
data.table.List method: Use the
.()function (which is an alias forlist()).## data.table way flights[, .(origin, dest)] ## Or flights[, list(origin, dest)] ## equivalent tidyverse way flights_tib %>% select(origin, dest)Character Vector Method: Use
c()with their character namesflights[, c("origin", "dest")]Range Method: Use
:to select variables within a range of columns.flights[, origin:dest]Prespecify Method: Define variables to keep outside of the
data.table, then usewith = FALSE. This option makes data.table not think thatkeep_vecis a varible in thedata.table.keep_vec <- c("origin", "dest") flights[, keep_vec, with = FALSE]To remove a column using the range or character methods, place a
!before the columns to removeflights[, !c("year", "month")] flights[, !(year:month)]To remove a column using the list method, assign that variable to be
NULLusing modify by reference (see below). If you run the below code, you’ll need to reload theflightsdata to get backyearandmonth.flights[, c("year", "month") := .(NULL, NULL)]Exercise: Use data.table to select the
year,month,day, andhourcolumns.Unlike the tidyverse, you filter rows and select columns in one call rather than using two separate functions.
## data.table way flights[origin == "JFK" & dest == "LAX", .(year, month, day, hour)] ## equivalent tidyverse way flights_tib %>% filter(origin == "JFK", dest == "LAX") %>% select(year, month, day, hour)Exercise: Use data.table to print out just the departure delays from JFK.
Creating New Variables (Mutate)
The fastest way to create and remove variables in a
data.tableis by reference, where we modify thedata.table, we don’t create a newdata.table.Use
:=to modify a data.table by reference. You put the variable names (as a character vector) to the left of:=. You put the new variables (as a list) to the right of:=.## data.table way flights[, c("gain") := .(dep_delay - arr_delay)] flights ## equivalent tidyverse way flights_tib %>% mutate(gain = dep_delay - arr_delay) -> flights_tib flights_tibQuickly remove a column by setting that variable to NULL.
## data.table way flights[, c("gain") := .(NULL)] flights ## equivalent tidyverse way flights_tib %>% select(-gain) -> flights_tib flights_tibAdd multiple variables by separating them with columns.
flights[, c("gain", "dist_km") := .(dep_delay - arr_delay, 1.61 * distance)] flights flights[, c("gain", "dist_km") := .(NULL, NULL)] flightsExercise: Add a variable called
speedthat is the average air speed of the plane in miles per hour. Then remove this variable.
Group Summaries
You calculate summaries in the column slot. It’s best to use the list method.
## data.table way flights[, .(dd = mean(dep_delay))] ## equivalent tidyverse way flights_tib %>% summarize(dd = mean(dep_delay))## data.table way flights[, .(dd = mean(dep_delay), ad = mean(arr_delay))] ## equivalent tidyverse way flights_tib %>% summarize(dd = mean(dep_delay), ad = mean(arr_delay))Count the number of rows with
.N.## data.table way flights[, .(.N)] ## equivalent tidyverse way flights_tib %>% count()In data.table, you create grouped summaries by simultaneously grouping and calculating summaries in one line, not separately like in dplyr.
In data.table, you use the
byargument to specify the grouping variable. It should also be a list.## data.table way flights[, .(dd = mean(dep_delay)) , by = .(origin)] ## equivalent tidyverse way flights_tib %>% group_by(origin) %>% summarize(dd = mean(dep_delay))You can calculate more than one group summary at a time, or group by more than one variable.
## data.table way flights[, .(dd = mean(dep_delay), .N), by = .(origin, carrier)] ## equivalent tidyverse way flights_tib %>% group_by(origin, carrier) %>% summarize(dd = mean(dep_delay), N = n())`summarise()` has grouped output by 'origin'. You can override using the `.groups` argument.Exercise: Use data.table to calculate the median air time for each month.
Exercise: Use data.table to calculate the number of trips from each airport for the carrier code DL.
Exercise: Use data.table to calculate the mean departure delay for each origin in the months of January and February.
Recoding
Suppose you want to change the values of a variable. In the tidyverse, we used
recode()to do this.In data.table, we filter the rows then mutate by reference.
Let’s substitute
24inhourto0.sort(unique(flights$hour)) sort(unique(flights_tib$hour))## data.table way flights[hour == 24L, hour := 0L] ## equivalent tidyverse way flights_tib %>% mutate(hour = recode(hour, `24` = 0L)) -> flights_tibsort(unique(flights$hour)) sort(unique(flights_tib$hour))Exercise: In the
originvariable, change"JFK"to"John F. Kennedy","LGA"to"LaGuardia", and"EWR"to"Newark Liberty".
Gathering
Problem: One variable spread across multiple columns.
Column names are actually values of a variable
Recall
table4aandtable4bfrom the tidyr packagedt4a <- as.data.table(tidyr::table4a) dt4b <- as.data.table(tidyr::table4b) dt4acountry 1999 2000 <char> <num> <num> 1: Afghanistan 745 2666 2: Brazil 37737 80488 3: China 212258 213766dt4bcountry 1999 2000 <char> <num> <num> 1: Afghanistan 1.999e+07 2.060e+07 2: Brazil 1.720e+08 1.745e+08 3: China 1.273e+09 1.280e+09Solution:
melt():## data.table way melt(dt4a, id.vars = c("country"), measure.vars = c("1999", "2000"), variable.name = "year", value.name = "count") ## Equivalent tidyverse way tidyr::table4a %>% gather(`1999`, `2000`, key = "year", value = "count") ## or tidyr::table4a %>% pivot_longer(cols = c("1999", "2000"), names_to = "year", values_to = "count")RDS visualization:
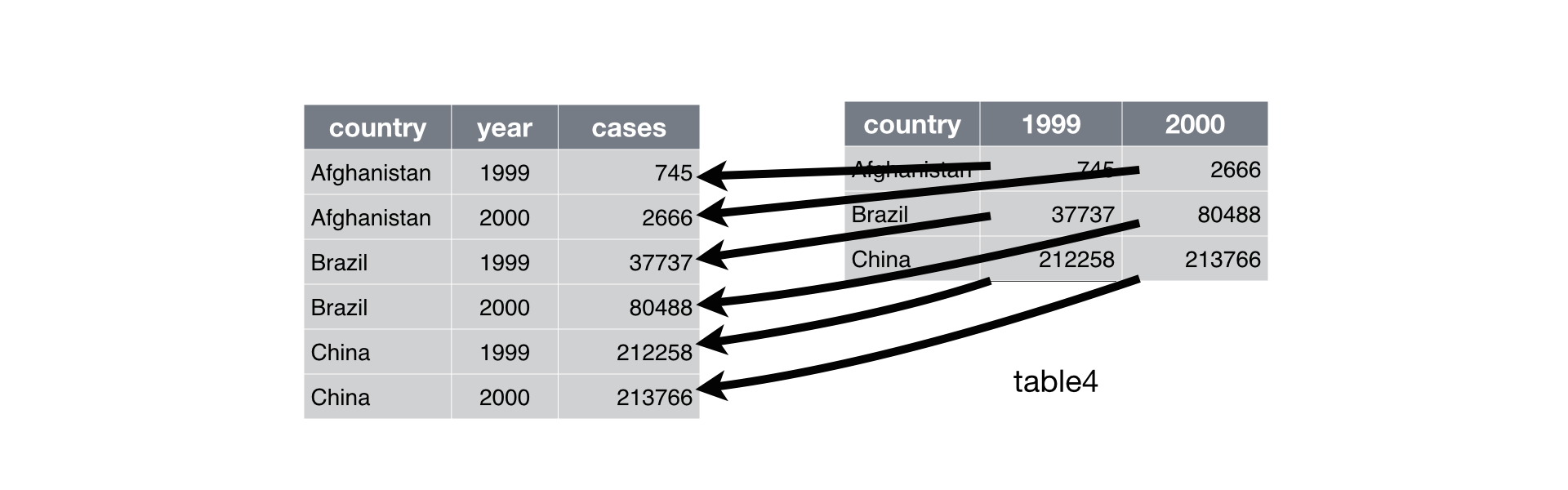
Exercise: gather the
monkeymemdata frame (available at https://data-science-master.github.io/lectures/data/tidy_exercise/monkeymem.csv). The cell values represent identification accuracy of some objects (in percent of 20 trials).
Spreading
Problem: One observation is spread across multiple rows.
One column contains variable names. One column contains values for the different variables.
Recall
table2from the tidyr packagedt2 <- as.data.table(tidyr::table2) dt2country year type count <char> <num> <char> <num> 1: Afghanistan 1999 cases 7.450e+02 2: Afghanistan 1999 population 1.999e+07 3: Afghanistan 2000 cases 2.666e+03 4: Afghanistan 2000 population 2.060e+07 5: Brazil 1999 cases 3.774e+04 6: Brazil 1999 population 1.720e+08 7: Brazil 2000 cases 8.049e+04 8: Brazil 2000 population 1.745e+08 9: China 1999 cases 2.123e+05 10: China 1999 population 1.273e+09 11: China 2000 cases 2.138e+05 12: China 2000 population 1.280e+09Solution:
dcast(). In theformulaargument, put the “id variables” to the left and the “key” variables to the right. In tidyverse jargon, thevalueis everything not stated in the formula and thekeyis everything to the left of the tilde.## data.table way dcast(dt2, formula = country + year ~ type, value.var = "count") ## Equivalent tidyverse way tidyr::table2 %>% spread(key = type, value = count) ## or tidyr::table2 %>% pivot_wider(id_cols = c("country", "year"), names_from = "type", values_from = "count")RDS visualization:
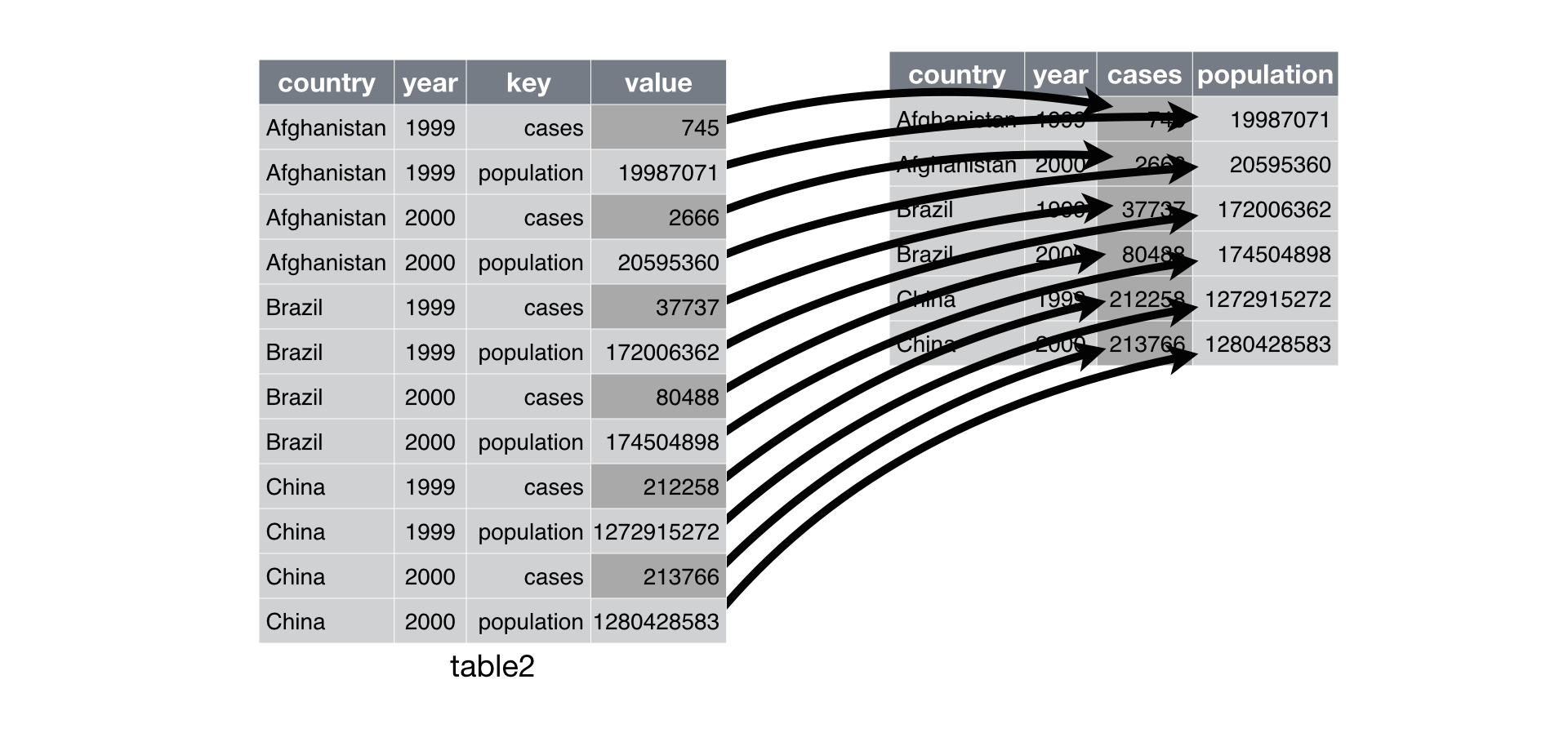
Exercise: Spread the
flowers1data frame (available at https://data-science-master.github.io/lectures/data/tidy_exercise/flowers1.csv).
Separating
To separate a column into two columns, use the base function
tstrsplit().dt3 <- as.data.table(tidyr::table3) dt3country year rate <char> <num> <char> 1: Afghanistan 1999 745/19987071 2: Afghanistan 2000 2666/20595360 3: Brazil 1999 37737/172006362 4: Brazil 2000 80488/174504898 5: China 1999 212258/1272915272 6: China 2000 213766/1280428583## data.table way dt3[, c("cases", "population") := tstrsplit(rate, split = "/")] dt3[, rate := NULL] dt3 ## equivalent tidyverse way tidyr::table3 %>% separate(rate, into = c("cases", "population"), sep = "/")RDS visualization:

Exercise: Separate the
flowers2data frame (available at https://data-science-master.github.io/lectures/data/tidy_exercise/flowers2.csv).
Uniting
To unite, use
paste().dt5 <- as.data.table(tidyr::table5) dt5country century year rate <char> <char> <char> <char> 1: Afghanistan 19 99 745/19987071 2: Afghanistan 20 00 2666/20595360 3: Brazil 19 99 37737/172006362 4: Brazil 20 00 80488/174504898 5: China 19 99 212258/1272915272 6: China 20 00 213766/1280428583## data.table way dt5[, year := paste(century, year, sep = "")] dt5[, century := NULL] dt5 ## Equivalent tidyverse way tidyr::table5 %>% unite(century, year, col = "year", sep = "")RDS visualization:

Exercise: Re-unite the data frame you separated from the flowers2 exercise. Use a comma for the separator.
Chaining
In the tidyverse, we chain commands by using the pipe
%>%. In data.table, we chain commands by adding additional brackets after the brackets we used. Data.table makes this very efficient.Let’s calculate the mean arrival delay for american airlines for each origin/destination pair, then order the results by origin in increasing order, breaking ties by destination in decreasing order.
## data.table way flights[carrier == "AA", .(ad = mean(arr_delay)), by = .(origin, dest)][order(origin, -dest)] ## Usual indentation for readability: flights[carrier == "AA", .(ad = mean(arr_delay)), by = .(origin, dest) ][order(origin, -dest)] ## Equivalent tidyverse way flights_tib %>% filter(carrier == "AA") %>% group_by(origin, dest) %>% summarize(ad = mean(arr_delay)) %>% arrange(origin, desc(dest))
Joining
We’ll use the following
data.tables to introduce joining.xdf <- data.table(mykey = c("1", "2", "3"), x_val = c("x1", "x2", "x3")) ydf <- data.table(mykey = c("1", "2", "4"), y_val = c("y1", "y2", "y3")) xdfmykey x_val <char> <char> 1: 1 x1 2: 2 x2 3: 3 x3ydfmykey y_val <char> <char> 1: 1 y1 2: 2 y2 3: 4 y3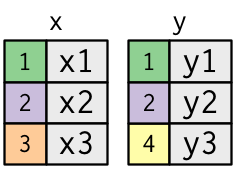
Use the
merge()function for all joining in data.table.Inner Join:
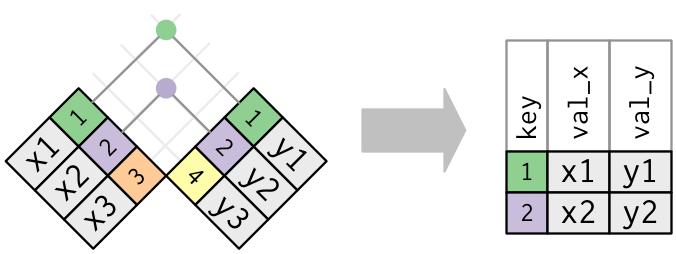
## data.table way merge(xdf, ydf, by = "mykey") ## equivalent tidyverse way inner_join(xdf, ydf, by = "mykey")Outer Joins
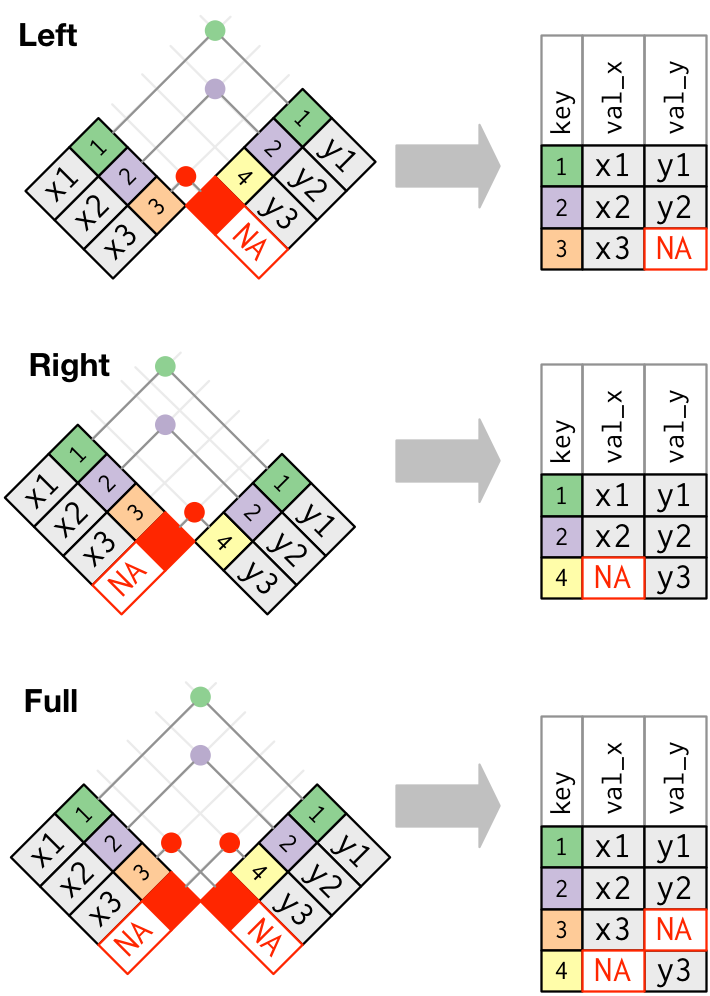
Left Join
## data.table way merge(xdf, ydf, by = "mykey", all.x = TRUE) ## equivalent tidyverse way left_join(xdf, ydf, by = "mykey")Right Join
## data.table way merge(xdf, ydf, by = "mykey", all.y = TRUE) ## equivalent tidyverse way right_join(xdf, ydf, by = "mykey")Outer Join
## data.table way merge(xdf, ydf, by = "mykey", all.x = TRUE, all.y = TRUE) ## equivalent tidyverse way full_join(xdf, ydf, by = "mykey")Binding Rows:
## data.table way rbind(xdf, ydf, fill = TRUE) ## equivalent tidyverse way bind_rows(xdf, ydf)When you have different key names, use the
by.xandby.yarguments instead of thebyargument.names(ydf)[1] <- "newkey" ydfnewkey y_val <char> <char> 1: 1 y1 2: 2 y2 3: 4 y3## data.table way merge(xdf, ydf, by.x = "mykey", by.y = "newkey") ## equivalent tidyverse way inner_join(xdf, ydf, by = c("mykey" = "newkey"))Exercise: Recall the
nycflights13datasetlibrary(nycflights13) data("flights") data("airlines") data("planes") flights <- as.data.table(flights) airlines <- as.data.table(airlines) planes <- as.data.table(planes)Add the full airline names to the
flightsdata.table.Exercise: Select all flights that use a plane where you have some annotation.
Additional References
There is so much more to data.table. You can do quite complicated and sophisticated data manipulations using data.table that would require a lot more typing in the tidyverse. I am not an expert in much of this functionality. A large number of articles and references are given on the homepage: https://rdatatable.gitlab.io/data.table/.