install.packages("reticulate")Python and Reticulate
Learning Objectives
- Set up R Studio environment for Python.
- Python REPL.
- Python chunks and Python scripts.
- Getting Started with Reticulate
- Installing Python Packages
Setting Up Python for R Studio
Most Python pros use Jupyter Notebook as their IDE when developing in Python.
But for R programmers who only need to dabble in Python (me, for example), we can use R Studio and the reticulate package.
R
You might need to accept the conda terms of service to use conda. So run the following code to load
reticulate.R
Sys.setenv(CONDA_PLUGINS_AUTO_ACCEPT_TOS = "yes") library(reticulate)You can also put this in your R environment (opened via
usethis::edit_r_environ()). Then you don’t need to set the enviornment variable every time you loadreticulate.CONDA_PLUGINS_AUTO_ACCEPT_TOS = "yes"To install a version of Python that reticulate will use, run in the terminal:
R
reticulate::install_miniconda()To validate that Python was installed and is available, run
R
reticulate::py_available()Python programmers typically modularize their projects into environments, which are separate installations of python and packages. That way, if you go back to a project, you go back to the exact same version of python and packages that you know work for that project. This reduces chances for breaking changes to affect old projects.
Use
conda_list()to list all of the environments.R
reticulate::conda_list()name 1 base 2 r-reticulate 3 anaconda3 4 msprime python 1 /home/dgerard/.local/share/r-miniconda/bin/python 2 /home/dgerard/.local/share/r-miniconda/envs/r-reticulate/bin/python 3 /home/dgerard/anaconda3/bin/python 4 /home/dgerard/anaconda3/envs/msprime/bin/pythonUse
conda_create()to create a new environment. You can choose the version of python that this environment uses.R
reticulate::conda_create(envname = "objects", python_version = "3.13.3")Use
use_condaenv()to specify the environment you want to use.R
reticulate::use_condaenv("objects")To install Python packages, use
py_install(). You need to specify the environment to which the packages are installed. For example, we can install the numpy, pandas, and matplotlib packages viaR
reticulate::py_install(envname = "objects", packages = c("numpy", "pandas", "matplotlib", "seaborn"))Note: We used conda environments here, but there is a competing system of environments called “virtualenv” which some folks use.
If you want to update the python version of a specific environment, I’ve found the easiest way was to just remove it with
conda_remove()and recreate it with the specified python version.
Python REPL’s, Chunks, and Scripts
To start an IPython shell — similar to the R command promp — run the following in R:
R
reticulate::repl_python()
“REPL” stands for “read–eval–print loop”, which is an interactive programming environment (like the R command prompt or the IPython shell).
You can exit the REPL by typing the following:
Python
exitYou can have Python chunks in R Markdown files by replacing the “
r” at the beginning of the chunk by “python”:```{python} # Code goes here ```You can access R objects in Python using the
robject. That is,r.xwill access, in Python, thexvariable defined using R.R
x <- c(1, 4, 6, 2)Python
r.x[1.0, 4.0, 6.0, 2.0]You can access Python objects in R using the
pyobject. That is,py$xwill access, in R, thexvariable defined using Pythong.Python
x = [8, 9, 11, 3]R
py$x[1] 8 9 11 3Sometimes it’s buggy, but you can usually begin a Python REPL by also hitting Control/Command + Enter inside the Python chunk:
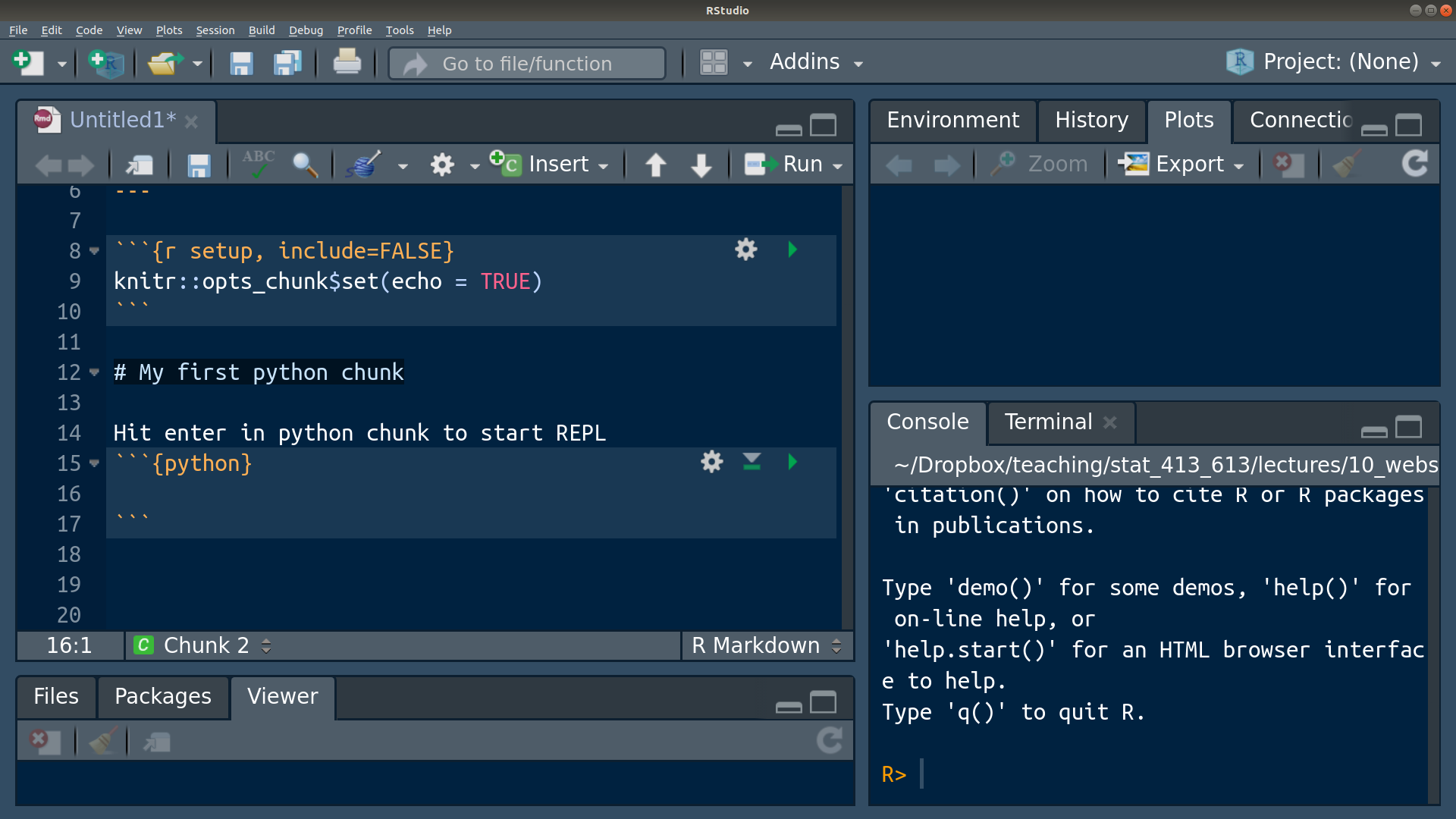
Python scripts (where there is only Python code and no plain text) end in “.py”. You can create a Python script in R Studio:
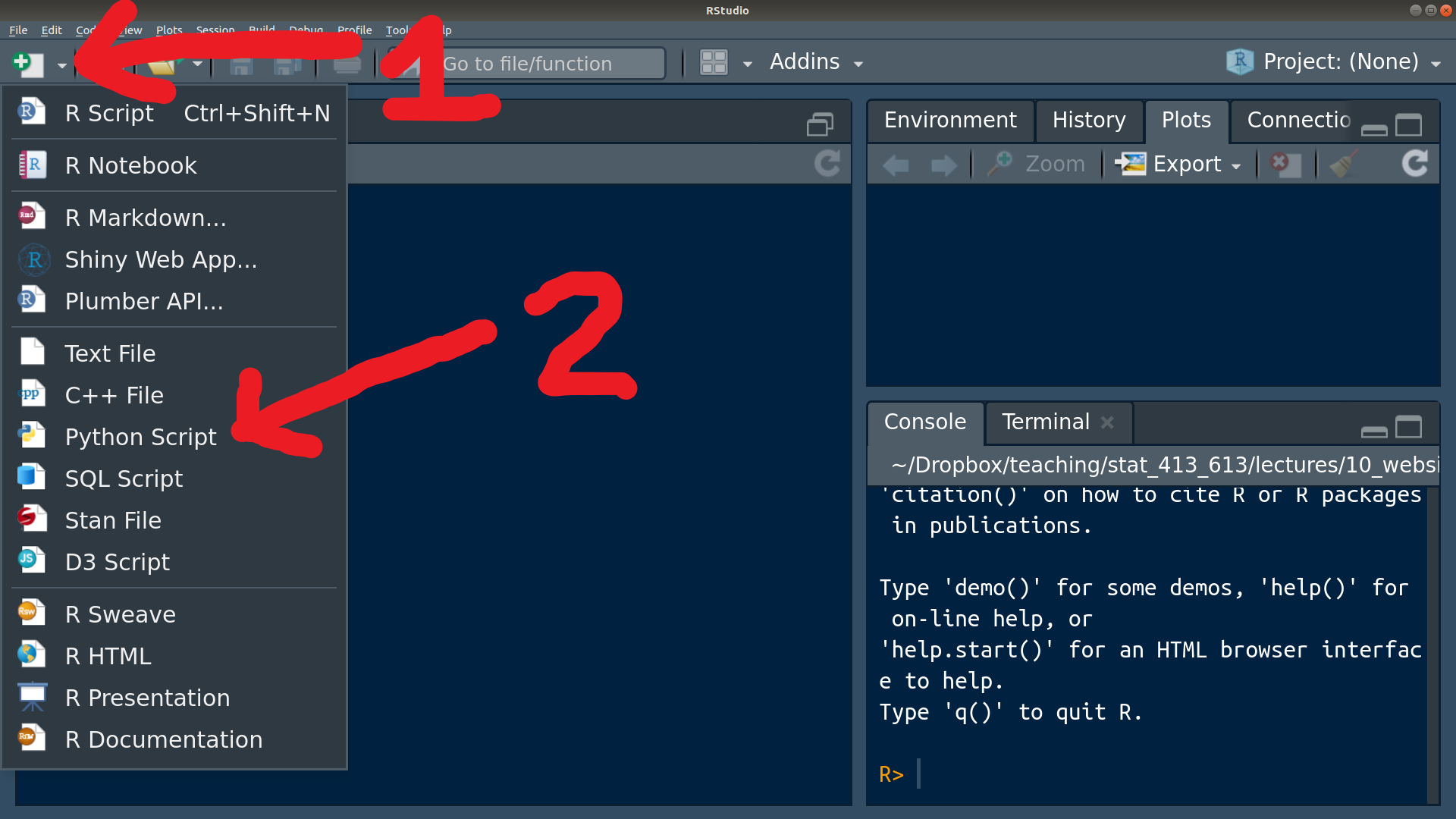
Hitting Control/Command + Enter inside a Python script will also start a Python REPL.